Note: This is the text of a 2014 Evo-Devo lecture by Eddy at the Pontifical Academy of Sciences at the Vatican. The slides corresponding to this presentation can be followed by opening the PowerPoint presentation immediately following this manuscript.
Evolving Concepts: The Role of Gene Loss from an Ancestral Genetic Tool-kit in Animal Evolution
Edward M. De Robertis
Introduction
In this meeting dedicated to evolving concepts of nature I would like to discuss some aspects of animal evolution. It is now clear from genome sequencing studies that bilateral animals evolved through the differential use of an ancestral “genomic tool-kit” shared by all bilateral animals. We will examine three main points (slide 1):
1) Whole-genome duplications followed by massive gene loss were very important evolutionary events.
2) Tandem duplications are accompanied by reciprocal gene deletions. Gene deletions are common and copious, but limit the potential for future evolutionary change.
3) Although a conserved set of genes is used by all animals, similar functions can be fulfilled by genes of different structure. In particular, we will examine the case of self-avoidance in neurons, which is mediated by different proteins in insects and humans.
1. Genomes contain the record of our evolutionary history
1.1 Developmental genes determine body shapes in animals
As explained by François Jacob (1982), “It is during embryonic development that the instructions contained in the genetic program of an organism are expressed, that the genotype is converted into phenotype. It is mainly the requirements of embryonic development that, among all possible changes in phenotype, screen the actual phenotypes”. Animals can be grouped into about 34 phyla, which have their cells arranged in such a way as to generate distinct body plans (slide 2). Of these, 30 constitute the bilateral animals which are most of the animals we see around us. These derive from a hypothetical common ancestor designated Urbilateria (Ur=primeval, bilateria=bilateral animal) that existed 560 million years ago (De Robertis and Sasai, 1996) (slide 3). One of the central problems in the field of evolution and development (Evo-Devo) is how morphologically complex this ancestral animal was.
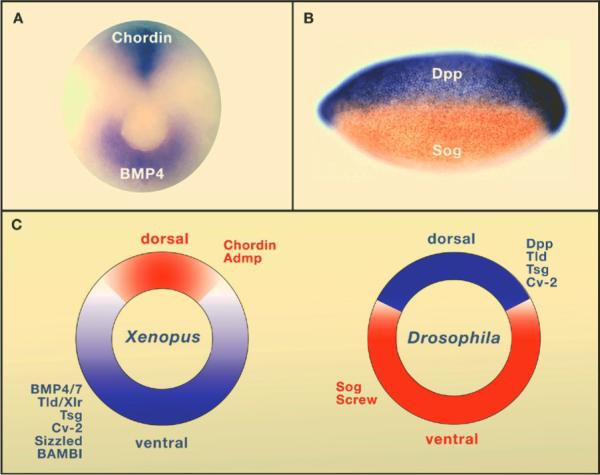
Genomes contain the record of our evolutionary history and we know that the vast majority of the genes used by animals today were present in Urbilateria and in its non-bilateral ancestors such as Cnidarians (e.g., sea anemones and Hydra). These genes are used to construct the protostome (mouth-first) animals which include most invertebrates, and the deuterostomes (mouth-second) which include chordates (such as the vertebrates, amphioxus and ascidians), hemichordates and echinoderms. Urbilateria patterned its body using conserved systems of developmental control genes such as the Chordin/BMP network that patterns Dorsal-Ventral (D-V) differentiation (slide 4) and the Hox genes that pattern the Antero-Posterior (A-P) axis.
1.2 D-V gene losses in mammals
The Chordin/BMP network patterns D-V differentiation in vertebrates, the fruit fly Drosophila, and many other animals. We have described a self-regulating network of secreted proteins that regulates gastrulation and is conserved in fishes, amphibians, reptiles and birds (De Robertis, 2008a) (slide 5). The platypus (Ornithorhynchus) is a mammalian ancestor that, surprisingly, has lost one of these genes, called ADMP, from its genome. Although platypus is a mammal, it still lays eggs in a nest. Although its reptile ancestors had two or three vitellogenin (yolk protein) genes, the platypus has retained only one (Warren et al. al., 2008). In higher mammals, two additional genes of this regulatory network, Crescent and Sizzled, were also deleted from the genome. Interestingly, in placental mammals (and also in marsupials) the vitellogenin gene was also completely lost. The Chordin/BMP network probably evolved to self-organize a morphogen gradient in cells actively migrating in a yolky egg. As soon as yolk disappeared, so did some genes in the network. Another possibility is that the lost genes were important in adjusting to differences in temperature during early development, which became unimportant when embryogenesis took place inside the mother. In a systematic search for genes lost in mammals but present in the chick genome, Kuraku and Kuratani (2011) found 147 losses of protein-coding genes. These losses of early patterning genes prompted our interest on the role of gene loss in evolution.
1.3 Gene losses in the Hox system
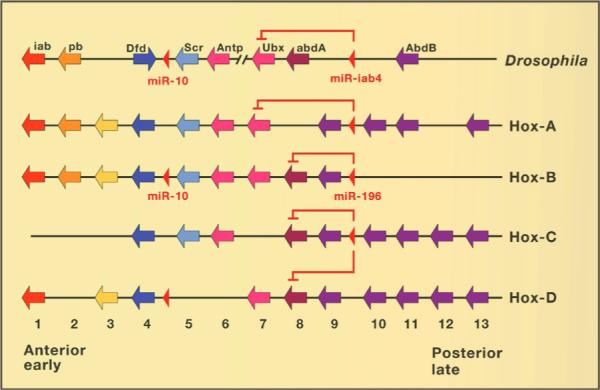
In Drosophila, homeotic genes specify A-P segment identity (slide 6). Edward B. Lewis found that there was a colinearity between their order in the DNA and the regions of the body they specify. Walter J. Gehring and others showed that all homeotic genes contained a conserved region called the homeodomain of 60 amino acids, which encodes a DNA-binding domain (Gehring, 1998). A Drosophila homeobox gene was used to isolate what constituted “the first development-controlling gene identified in vertebrates” thirty years ago (Carrasco et al., 1984) (slide 7). Hox gene complexes have been isolated through the work of many groups and found to conserve the colinearity in Drosophila and chordates (slide 8). From these studies, one can conclude that a Hox complex consisting of at least six genes was present in Urbilateria (De Robertis, 2008a; Holland, 2013). Mice and men have four Hox complexes that represent duplications from an ancestral gene set consisting of 13 paralogues (duplicated genes). Within these complexes 13 individual genes have been lost, illustrating that genes are not only gained but also readily lost.
As discussed in section 2.3, sequencing of the genome of the chordate ancestor amphioxus (Branchiostoma) has proven particularly informative. It contains a single Hox complex (slide 8), which indicates that the vertebrates underwent two whole-genome duplications during the early evolution of a fish ancestor (slide 9).
2. Evolution by whole genome duplication followed by gene loss
2.1 Susumu Ohno and evolution by gene duplication
In 1970 Susumo Ohno published a prescient book entitled “Evolution by gene duplication” (slide 10). After looking at chromosomes and quantifying DNA contents in nuclei after staining DNA with Feuglen reagent, he predicted that two whole-genome duplications events (4X) had occurred early during vertebrate evolution.
These duplications probably were key to the evolutionary success of the vertebrates. Ohno also predicted that bony fishes (teleosts) had undergone an additional round of genome duplications (8X) when compared to their cartilaginous ancestors. In fact, fishes are the most successful of all vertebrate animals in terms of having the most species and number of individuals. He also noted that the surf smelt (8X) had half the genome size of the trout (16X), and the salmon (32X) had double that of the trout. Therefore, these familiar fish constitute a polyploid series of nutritious animals. Polyploidization events have occurred multiple time in fishes and amphibians, but ceased in reptiles, birds and mammals, when sex chromosomes became distinct. The role of gene duplication – both by whole genome and by tandem DNA duplication – in providing a substratum for evolutionary innovation is now well recognized. When two genes are duplicated one of the copies can adopt different functions or a different regulation. The role of gene deletion is less generally recognized.
2.2. Tandem duplications are accompanied by deletions
When two chromosomes undergo crossing over in meiosis (or sister chromatid exchange in germ line mitoses) they usually align exactly. However, if the two DNA strands are not aligned correctly (unequal crossing over) this will result in a tandem duplication of one DNA strand. What is less frequently realized is that for each duplication there is an accompanying reciprocal deletion in the other DNA molecule (slide 11).
Deletions by unequal crossing over must be very frequent, because they provide the molecular mechanism by which deleterious sequence changes are eliminated in repeated genes. For example, the Xenopus laevis nucleolar organizer has 450 tandemly repeated large ribosomal RNA (rDNA) genes (Brown and Dawid, 1968). Mutations must occur at random in rDNA, just as in any other DNA sequence. To keep the 450 rDNA genes homogeneous in sequence, constant deletions accompanied by new tandem duplications are required. Through these continuous gene deletions, the nucleolar organizer can cleanse itself of genes that have become inactivated by mutation. In Drosophila, there are normally about 100 rDNA copies. The bobbed mutant has an insufficient number of rDNA copies for vigorous growth, resulting in small flies. However, the bobbed mutation can easily revert by unequal crossing over, resulting in an increase in rDNA genes and normal growth. In this way, the rDNA gene sequences are maintained invariant by continuous deletion and duplication events mediated by unequal crossing over (Ritossa et al., 1966; Ohno, 1970).
These examples argue that gene loss must be copious and easily achieved though unequal crossing over events that generate deletions and duplications. Natural selection (Darwin, 1858) provides the constant policing required to ensure that all genes required for propagation of the species with maximal reproductive fitness are preserved. All animals produce many more progeny than the numbers needed to maintain their population. An extreme example is provided by the Echinus esculentus sea urchin female, which releases 20 million eggs, even though only two surviving adult progeny are required to maintain the same number of individuals in the population. The price of gene loss is that many offspring will not be viable, but that is a small price to pay for variation when reproduction is so exuberant.
Genomes are probably constantly being streamlined by removal of unwanted DNA. As computer programmers know, as a large program grows over time the software becomes unwieldy because many layers of no longer useful code remain. Some programmers specialize in locating these unnecessary instructions and deleting them. In the case of the genome, DNA deletions are constantly purifying genomes of unwanted DNA code. Gene loss can serve the useful function of increasing fitness.
2.3 The amphioxus and yeast genomes have been very informative
Sequencing of the amphioxus genome confirmed Ohno’s proposal that vertebrates had undergone two whole-genome duplication events. Amphioxus is an early chordate that did not undergo genome duplication. In principle, humans should have four copies of every chordate gene. In the case of the Hox gene complexes, all four copies have been preserved. Comparison of the amphioxus and human genomes showed that 25% of human genes have two or more copies resulting from the two genome duplications (Putnam et al., 2008) (slide 12). Massive gene losses, through the process known as rediploidization, have left humans with 75% of their protein-coding genes as single copies. Developmental control genes appear to have been conserved in duplicated form more frequently than housekeeping genes.
Comparison of the amphioxus and human genomes has shown that the tunicate class of chordates has undergone massive gene losses (slide 13). These animals, also known as ascidians or sea squirts, have a larval swimming form with a notochord but then adopt a sessile life form in which they attach to rocks and become essentially a large water-filtering pharynx. Ascidians have not undergone whole genome-duplications and would be expected to have a similar number of genes as amphioxus. However, the ascidian genome has lost a total of 2,250 – out of a total of about 8,000 – gene families that are present both in amphioxus and humans (Holland et al., 2008). Adaptation to this particular life form came at the cost of future evolution for these animals.
The baker’s yeast Saccharomyces cerevisiae evolved from a single whole-genome duplication followed by massive gene losses (Wolfe and Shields, 1997). Sequencing of a diploid ancestral yeast (Kluyveromyces waltii) species revealed that a tetrapoidization was followed by massive gene losses through gene deletions (Kellis et al., 2004) (slide 14). In fact, one copy of each duplicated gene has been deleted for 88% of all S. cerevisiae gene loci through a process involving deletions comprising multiple adjacent genes (Kellis et al., 2004). In present-day S. cerevisiae only 12% of the genes have been maintained in duplicated form. Frequently, the protein encoded by one of the duplicated genes has diverged significantly in sequence from the ancestral gene pair. The yeast genome has provided very strong support for the hypothesis that gene duplication allows one of the duplicated copies to become different in function (Ohno, 1970). Genome duplication is thus followed by rapid loss of many, but not all, genes.
3. Loss of genes and anatomical structures in evolution
3.1 Losses by disuse
The old adage that “what is not used atrophies” was recognized from the time of French naturalist Buffon. When a change in habits or in the environment occurs, it seems likely that all variations that are compatible with the new behavior will be allowed to escape the strictures of Darwinian natural selection. There are many examples of this.
The case of the gastrointestinal tract of the vampire bat provides a striking illustration of adaptations that follow a change in feeding habits (slide 15). The external morphology of the vampires is very similar to that of the insectivorous bats from which they evolved. However, the esophagus has become very thin, with a lumen so narrow that only liquids can pass (Kent, 1973). In addition, the stomach has been modified to provide a lateral sac for the accumulation and dehydration of blood. These gastrointestinal adaptations have rendered vampires unable to feed on insects as the other bats do. These internal adaptations are related to the change in feeding habit and likely occurred in a relatively short evolutionary period because vampires are very similar in external morphology to their close insectivorous relatives. These variations would also occur spontaneously in insectivorous bat species, but the guiding hand of natural selection would have prevented them from appearing in the population for they would not have been able to swallow insects.
Laboratory mice have been bred in captivity for centuries, starting by the breeding of “fancy” mice pets in China and then Japan. Unlike field mice which reproduce seasonally, under domestication the artificial selection imposed by human breeders has favored reproduction throughout the year. The change was caused by loss-of-function mutations in two genes which encode enzymes required for the conversion of serotonin into melatonin in the pineal gland (Olson, 1999). Melatonin secretion normally monitors the length of daylight hours, adjusting reproduction to the seasons. In addition, most laboratory strains of mice are deaf, which indicates another loss-of-function.
Darwin himself dealt with the effects of disuse in his Origin of Species. He noted that beetles in windswept islands become flightless (200 out of the 550 beetle species in the island of Madeira). Darwin also discussed other examples of adaptive organ losses such as flightless birds and moles with rudimentary eyes (Darwin, 1859).
When a change in habits or in the environment occurs, it seems likely that all variations that are compatible with the new behavior will be allowed to escape natural selection, with adaptations secondary to a change in life habits following. These adaptations will become fixed when new speciation events occur.
3.2 DNA deletions and the adaptation of fish to new environments
An important question is which genes were actually mutated during evolution in nature. Three-spined sticklebacks are marine fish that spawn in fresh water. They became entrapped independently in many lakes after the last ice age. Only 10,000 to 20,000 years ago Canada and the great lakes were covered by the Laurentide ice sheet. As the glaciers receded by global warming, lakes were formed and the local stickleback populations diversified rapidly and dramatically (slide 16). These events were recent enough that genetic crosses can still be carried out. Isolation can cause variations that constrain reproduction (such as different mating dances), but fertile progeny will result if sperm and eggs are squeezed into a Petri dish. This allows genes responsible for morphological adaptations to be mapped genetically (Jones et al., 2012).
Sticklebacks have spines that prevent predators from swallowing them. They have two prominent pelvic spines that have been lost repeatedly in different lakes (slide 17). Why would sticklebacks ever lose the pelvis? Shallow lakes often contain dragonfly nymphs that grasp small fish and eat them. The pelvic spines greatly facilitate the grasping predators. Genetic mapping and sequencing has shown that the mutation is due to the deletion of a DNA enhancer element that is required for the expression of a gene called Pitx1 specifically in the pelvis (Chan et al., 2010) (slide 18). Enhancer elements direct the expression of genes to specific tissues. Deletion of enhancer DNA has the advantage that the Pitx1 protein gene, which is also expressed in other regions, is left in place. Loss of the Pitx1 protein coding region would otherwise be lethal. The extraordinary finding that three different deletions occurred independently of each other, always removing the same gene enhancer. Adaptation by gene loss must be frequent as it occurred repeatedly in a short time from an evolutionary perspective.
Sticklebacks also protect themselves with armored plates, which have been lost in many populations (Jones et al., 2012). This mutation is caused by deletion of the enhancer for a growth factor gene called ectodysplasin that induces the formation of bony plates in the skin. Unlike the previous case, the deletion of the enhancer was present in the marine population before sticklebacks became isolated at the end of the ice age. Thus, all populations carry the same mutation. It does not manifest itself in the large marine population because it is a rare recessive trait. As the armor is probably energetically expensive, if not needed in the absence of predators, this mutation was naturally selected for in some of the isolated lake populations.
Loss-of-function mutations can also cause deletions in the protein coding genes themselves in addition to their enhancer elements. Many cave animals, such as shrimp, salamanders and fish lose pigmentation and eyes when they adapt to permanent darkness. Mexican Tetra river fish became entrapped in subterranean caves multiple independent times. Genetic crosses have mapped their albinism to Oca2. The ocular and cutaneous albinism-2 protein encodes a Tyrosine transporter; Tyrosine is the amino acid building block for melanin. Oca2 encodes a very large gene which appears not to have many pleiotropic effects and its mutation is the most frequent cause of albinism in humans. Interestingly, independent deletions in the Oca2 genes occurred repeatedly in various Tetra cave populations (Protas et al., 2006; Rohner et al., 2013). Variation in evolution seems to follow the path of least resistance. The ancestral genetic tool-kit has a profound influence, channeling the type of variations that result in adaptations.
Humans carry enormous numbers of recessive mutations that could serve as a reservoir of variation in case of a drastic change in the environment. Sequencing of human genomes indicates that each of us carries about 100 mutations that would inactivate protein-coding genes, usually in heterozygote form, with about 20 protein genes having been deleted (MacArthur et al., 2012).
4. Urbilateria: a complex or simple animal
4.1 Urbilateria had a complex life cycle
A central question in Evo-Devo is how complex the urbilaterian ancestor was from a morphological point of view. From a genetic point of view it must have contained the vast majority of the genes present in bilateral animals today. Urbilateria probably had a complex life cycle with an initial ciliated free-swimming (pelagic) larval phase in oceanic plankton (slide 19). Many phyla of the two main branches of bilateral animals (e.g., annelids, molluscs, hemichordates and echinoderms) have planktonic larvae with two ciliary bands that collect food into the mouth by beating in opposite directions, an apical eye, and an apical ciliary tuft. These larvae then settle on the ocean bottom for their adult life (benthic) phase. The ancestral planktonic phase of the life cycle has been lost repeatedly in many marine animal lineages.
Much debate has centered on whether Urbilateria was segmented (De Robertis, 2008b). The segment formation mechanism in Drosophila is very different from that of the vertebrates. In vertebrates, segmentation is caused by an oscillating network of genes of the Notch pathway (Deqéant et al., 2006). Studies on the segmentation of the cockroach embryo have also shown oscillatory expression of Notch pathway genes, indicating that this might be the ancestral mode of segmentation (Pueyo et al., 2008). The ability to form segments may have been lost secondarily in non-segmented bilateral animals, such as planarians and nematodes. The alternative view is that the ancestor was a simple organism such as a planarian and metamerism arose multiple times in evolution.
4.2 The axochord as a homolog of the notochord
The chordate phylum is characterized by a dorsal flexible rod that is used for undulatory swimming and as a signaling center in the embryo (slide 20). In the vertebrates the notochord serves as an induction center for the formation of the vertebrae, and later degenerates remaining only as the nucleus pulposus of the intervertebral disc. In a recent study Detlev Arendt and colleagues discovered that the ventral midline of annelid embryos has a medial muscle called the axochord (slide 21). They found that the axochord muscle expresses many genes similar to those found in the vertebrate notochord. These include homologs of Brachyury, Twist, FoxD, Fox1, Collagen A, Netrin, Slit, Noggin and Hedgehog (Lauri et al., 2014). The combination of such genes in one anatomical structure is found only in the axochord and notochord. In annelids, mollusks and chaetognaths, a contractile muscular axochord-like organ serves for the attachment of oblique muscles to its collagenous sheath. In Drosophila, the muscular nature was lost, but the signaling activity was retained by the mesodermal midline glia cells.
In amphioxus, the notochord became more rigid through a process of intracellular vacuolization that turned the notochord into a rod of fixed length (slide 22). Interestingly, amphioxus notochord cells still have a muscular nature. Contraction of the amphioxus notochord serves to regulate the stiffness of this rod essential for undulatory swimming (which in amphioxus can take place both in forward and reverse directions; Webb, 1973). These notochord muscle fibers were lost in the vertebrates. In mammals, the vacuolization of the notochord was lost and it remained as a very thin structure serving as a signaling center in development but which is no longer used in locomotion. Thus, it appears that the urbilaterian ancestor had a muscular mesodermal midline that then became vacuolized, increased its collagenous nature, and finally served as an inducing center for the formation of the vertebral column.
5. Using a common tool-kit in different ways can generate diversity
5.1 Conserved and novel functions in the central nervous system (CNS)
There are many examples of genes that have highly conserved functions in the CNS of vertebrates and invertebrates (De Robertis, 2008a). The most celebrated case of gene conservation is Pax6, the master control gene used for eye development in all animals (Gehring, 1998).
While a conserved gene tool-kit is shared by animals, genes are not always used in the same way. As Pontifical Academy President Werner Arber has said: “Nature is very inventive and able to find different ways to reach a specific goal”. One of the best examples of this incredible versatility is provided by the molecular solution to the problem of neuronal self-avoidance (slide 23).
Self-avoidance is an essential property of neurons by which dendrites and axons from the same neuron never contact each other. Projections are repelled if they contact a membrane from the same cell. There are 100 billion neurons in the human CNS and each one has its own identification tag. This may be considered similar to the license plates of cars, which gives them uniqueness. Self-avoidance is essential for neurons to form proper circuits. For example, it ensures that a neuron will cover an entire area without making contact with its sister branches. In Drosophila this problem has been resolved by the elaboration of a single, remarkable gene.
Work by the laboratory of Larry Zipursky at UCLA has shown that mutation of Dscam1 (Down syndrome cell adhesion molecule-1) eliminates repulsion between branches of the same neuron (slide 24). Dscam1 evolved multiple exon sequences for three of its nine immunoglobulin domains. These segments are then spliced together at the mRNA level to encode a transmembrane protein. The three variable exons have 12, 48 and 33 different possibilities respectively, as well as 2 different transmembrane domains that target Dscam1 to axons and dendrites (Zipursky and Grueber, 2013). Alternative splicing from this one gene can generate a staggering number of different proteins: 12 x 48 x 33 x 2 = 38,016, all from one gene (slide 25). This is an amazing number of proteins, considering that the entire Drosophila genome encodes a total of 15,000 (and the human one about 20,000) different protein-coding genes.
Self-avoidance occurs only when the three variable domains of Dscam1 match each other exactly. When two Dscam1 proteins bind homotypically in different membranes, this triggers repulsion via the intracellular domain and the cytoskeleton machinery, and the two sister cell projections separate. Each neuron expresses about 10-20 different Dscam1 isoforms in a stochastic fashion, and this provides an enormous amount of combinatorial possibilities for each individual neuron (Miura et al., 2013).
Remarkably, humans use a different mechanism for neuronal self- avoidance. We do have two Dscam genes, but they lack all the alternative exons found in insects and therefore encode only one protein each. In addition, Dscam is also not variable in the annelids. This suggests that Urbilateria had an ancestral Dscam gene that was adapted specifically in the insect lineage to generate the self-avoidance system.
Recent work by Tom Maniatis and Josh Sanes has shown that in mammals the self-avoidance function is fulfilled by an entirely different gene family called the clustered protocadherins (slide 26). Rather than using splicing to generate variability, protocadherins utilize alternative promoters that result in the splicing of different extracellular domains to a common intracellular region (Lefebvre et al., 2012). Several protocadherins are expressed in a stochastic fashion in neurons. They also have highly specific homophilic binding. Although there are only 50 different protocadherin genes, these proteins form hetero-tetramers which greatly increases the combined variability. As in the case of Dscam1, homophilic binding triggers repulsion and self-avoidance (Thu et al., 2014). Protocadherins are very different transmembrane proteins from Dscam. Homotypic recognition occurs through cadherin domains instead of immunoglobulin domains. However, the intracellular region also has the capacity of transmitting to the cytoskeleton a message of repulsion.
Surprisingly, insect genomes do not contain a single protocadherin gene. However, spiders do have one protocadherin. This indicates that Urbilateria, and perhaps its ancestors, already had a protocadherin gene containing six cadherin domains. Different solutions to the physiological problem of self-avoidance have been achieved through the gain of variability in different proteins. The resourcefulness of evolution is enormous.
6. Chance and Necessity in evolution
Darwinian evolution has been used many times as an argument against Judeo-Christian faith. In a very interesting book entitled “In the Beginning…” Cardinal Joseph Ratzinger of Munich (later member of our Academy and Pope Benedict XVI), wrote: “There has been a conflict between natural sciences and theology that has been a burden to the Church. This did not have to be”. This book contains the four Lent homilies Ratzinger gave in the Frauenkirche Cathedral in response to the highly influential book “Chance and Necessity” written in 1970 by the famous French geneticist Jacques Monod (slide 27). The Cardinal’s audience must have been very faithful and patient to sit through this thoughtful argument on Evolution and Faith in the course of four weeks (in fact, he gratefully dedicated his book to them).
Monod’s essay on the natural philosophy of modern biology proposed a new “ethics of knowledge”. The main guiding principle was the principle of objectivity (in essence similar to an earlier proposal by physiologist Claude Bernard) by which hypotheses must be tested by experiment and the results analyzed by reason, and not influenced by any other values or ideologies. His essay, however, was influenced by atheism and his book ended in a dark philosophical message: “The ancient covenant is in pieces; man knows at last that he is alone in the universe’s unfeeling immensity, out of which he emerged only by chance.”
François Jacob was the close collaborator of Monod in their Nobel prize-winning work on gene regulation. He took a more optimistic view in his own book on evolution: “Western science is founded on the monastic doctrine of an orderly universe founded by God who stands outside of nature and controls it through laws accessible to human reason” (Jacob, 1982). The response from Cardinal Ratzinger was related, making the argument that God is also Reason and set up universal laws by the act of Creation. A similar reflection was made by Pope Francis in the Domus Sanctae Marthae homily cited in the introduction to this meeting on Evolving Concepts of Nature (slide 28): “God made things – each one – and he let them go with the interior, inward laws which he gave to each one, so that they would develop, so they would reach fullness.”
Cardinal Ratzinger explains at the end of “In the Beginning…” why this is an important issue to the Catholic Church: “Only if the Redeemer is also Creator can he really be Redeemer. That is why the question of what we do is decided by the ground of what we are. We can win the future only if we do not lose creation.” For Ratzinger a Creator that set the rational natural world is essential; otherwise, he could not have sent his Son to save us.
The fields of religion and science are separate. The scientific mind would argue that invoking a Creator because we need salvation is a circular argument. It indeed is, but for Christians it is clear how much good Faith can do in the daily lives of so many people. Conversely, in the field of science we know that a natural world from which universal laws can be uncovered by experiment must clearly exist, because of the beneficial effects that scientific development has brought in the improvement of human life.
As pointed by Monod, the principle of science is that we must deduce the laws of the natural world with complete objectivity, not influenced by biases or ideologies. It was in the Church that the separation between the pagan (e.g., Greek philosophy) and the sacred was first made. In the natural sciences hypotheses are tested by experiments interpreted by our own human reason. It is therefore immaterial from the point of view of the advancement of everyday science whether the first principle is Chance as in Monod’s view, or through a Creator as in the Judeo-Christian view. All that is needed is a rational universe governed by natural laws.
7. Conclusions
7.1 Evolving concepts of evolution
Work in developmental genetics has revealed that animal evolution is generated through variations in ancestral gene networks that were present in Urbilateria, the common ancestor of all bilateral animals and in its metazoan ancestors (slide 30). Animal morphologies depend on the arrangement of cells with respect to each other and this is controlled by ancient developmental gene networks. The discovery of conserved Hox genes thirty years ago, revealed that the mechanisms of development were common to all animals. The use of developmental control networks placed evolutionary constraints on the types of animal anatomies that can evolve by natural selection. The “Necessity” aspect of animal evolution resides on the conserved use of genes of the machinery of embryonic development. Not all possible body plans can evolve; only those that are compatible with the developmental gene networks, which are in turn determined by our previous history.
An important source of variation in evolution is through duplications and deletions of this ancient set of genes. The urbilaterian genome must have had a surprisingly complete set of the genes used in all animals (slide 30). Tandem gene duplications provided a way of generating new genes that can then adopt new functions or be expressed in specialized regions. Each tandem duplication is accompanied by a reciprocal deletion, which plays a fundamental role in policing the genome. Natural selection provides the constant vigilance required to ensure that all genes required for propagation of the species with maximal reproductive fitness are preserved.
Genome-wide duplications provided a very effective way of providing new genes that can then diverge in function. Whole-genome duplications occurred many times in plants, yeasts, and early vertebrates. Two successive genome-wide duplications were key to the evolutionary success of vertebrates. These events were followed by massive gene loss of genes that did not acquire a specialized function. If there were no gene losses all human genes should be present in four copies, but in fact 75% of them are now single-copy.
The adaptive role of gene loss in evolution has not been fully appreciated. When the environment changes, losses of DNA enhancer elements or of protein-coding regions have been documented in stickleback and cave fishes. In lakes without predators, sticklebacks rapidly lose their armor, using a deletion mutation that exists at low frequency in the large marine population. Extra features such as body armor come at a fitness cost. Once there are no predators, it becomes adaptive to lose the unneeded feature. Cave fish lose their eyes because the energetic cost of vision is high; vision consumes more energy in the dark. Gene losses can result in better adaptation, but at the same time limit the potential for future evolution. The bilateral animal common ancestor started this lineage with an enormous reservoir of genetic diversity which was trimmed and elaborated by natural selection of the fittest, in some lineages more than in others. Life on earth evolved though a series of unique events such as the establishment of single genetic code, of the cell membrane, of photosynthesis, of multicellular animals and of a common tool-kit for bilateral animals.
On other side of the coin, very different genes can be used to achieve the same physiological needs. We have reviewed this in the case of self-avoidance in neurons, which is mediated by alternatively spliced Dscam1 insects and by tandemly repeated protocadherins in humans. Although different, both proteins used the principle of homotypic recognition to trigger repulsion at the level of the cell cytoskeleton.
In sum, bilateral animals evolved through variations in ancestral developmental gene networks in their DNA. It seems a miracle that we humans got to where we are today through evolutionary gains and losses of an ancient set of genes.
Bibliography
Brown, D.D. and Dawid, I.B. (1968), specific gene amplification in oocytes. Science 160, 272-280.
Carrasco, A.E., McGinnis, W., Gehring, W.J. and De Robertis, E.M. (1984), Cloning of a Xenopus laevis gene expressed during early embryogenesis that codes for a peptide region homologous to Drosophila homeotic genes: implications for vertebrate development. Cell 37, 409-414.
Chan, Y.F. et al. (2010). Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327, 302-305.
Darwin, C. (1859), On the Origin of Species by Means of Natural Selection, or Preservation of Favored Races in the Struggle for Life, Murray, London
Deqéant, M.L. et al. (2006), A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science 314, 1595-1598.
De Robertis, E.M. (2008a), Evo-Devo: Variations on ancestral themes. Cell 132, 185-195.
De Robertis, E.M. (2008b), The molecular ancestry of segmentation mechanisms. Proc. Nat. Acad. Sci. USA 105, 16411-16412.
De Robertis, E.M. and Sasai, Y. (1996), A common plan for dorso-ventral patterning in Bilateria. Nature 380, 37-40.
Gehring, W.J. (1998), Master Control Genes in Development and Evolution: The Homeobox Story, Yale Univ. Press, New Haven.
Holland, P. (2013). Evolution of homeobox genes. WIREs Dev. Biol. 31, 31-45.
Holland, L.Z. (2008), The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Research 18, 1100-1111.
Jacob, F. (1982). The Possible and the Actual, University of Washington Press, Seattle.
Jones, F.C. et al. (2012). The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484, 55-61.
Kellis, M., Birren, B.W., and Lander, E.S. (2004). Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428, 617-624.
Kent, G.C. (1973), Comparative anatomy of the vertebrates. The C. V. Mosby Company, Saint Louis.
Kuraku, S., and Kuratani, S. (2011), Genome-wide detection of gene extinction in early mammalian evolution. Genome Bio. Evol. 3, 1449-1462.
Lauri, A. et. al. (2014), Development of the annelid axochord: Insights into notochord evolution. Science 345, 1365-1368.
Lefebvre, J.L., Kostadinov, D., Chen, W.V., Maniatis, T., and Sanes, J.R. (2012), Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature 488, 517-521.
MacArthur, D.G. et al. (2012) A systematic survey of loss-of-function variants in human protein-coding genes. Science 335, 823-828.
Miura, S.K., Martins, A., Zhang, K.X., Graveley, B.R. and Zipursky, S.L. (2013). Probabilistic splicing of Dscam1 establishes identity at the level of single neurons. Cell 155, 1166-1177.
Monod, J. (1971). Chance & Neccesity, Alfred A. Knopf, New York.
Ohno, S. (1970), Evolution by Gene Duplication, Springer-Verlag, Heidelberg.
Olson, M.V. (1999), Molecular Evolution ’99. When less is more: gene loss as an engine of evolutionary change. Am. J. Hum. Genet. 64, 18-23.
Protas, M.E., Hersey, C., Kochanek, D., Zhous, Y., Wilkens, H., Jeffery, W.R., Zon, L.I., Borowsky, R. and Tabin, C.J. (2006), Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism, Nat. Gen. 38, 107-111.
Pueyo, J.I., Lanfear, R., Couseo, J.P. (2008), Ancestral notch-mediated segmentation revealed in the cockroach Periplaneta Americana. Proc. Natl. Acad. Sci. USA 105, 16614-16619.
Putnam, N.H. et al. (2008), The amphioxus genome and the evolution of the chordate karyotype. Nature 453, 1064-1071.
Ratzinger, J. (1995), In the Beginning, B. Eerdmans Publishing Co., Michigan.
Ritossa, F.M., Atwood, K.C. and Spegelman, S. (1966). A molecular explanation of the bobbed mutants of Drosophila as partial deficiencies of “ribosomal” DNA. Genetics 54, 819-834.
Rohner, N., Jarosz, D.F., Kowalko, J.E., Yoshizawa, M., Jeffery, W.R., Borowsky, R.L., Linquist, S. and Tabin, C.J. (2013), Cryptic variation in morphological evolution: HSP90 as a capacitor for loss of eyes in cavefish. Science 342, 1372-1375.
Thu, C.A. et al. (2014), Single-cell identity generated by combinatorial homophilic interactions between α, β, and γ protocadherins. Cell 158, 1045-1059
Warren, W.C. (2008), Genome analysis of the platypus reveals unique signatures of evolution. Nature 453, 175-183.
Webb, J.E. (1973), The role of the notochord in forward and reverse swimming and burrowing in the amphioxus Branchiostoma lanceolatum. J. Zool. 170, 325-338.
Wolfe, K.H., and Shields, D.C. (1997). Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387, 708-713.
Zipursky, S.L. and Grueber, W.B. (2013), The molecular basis of self-avoidance. Annu. Rev. Neurosci 36, 547-568.
| The Powerpoint presentation for this talk can be accessed by opening the file beside this text. |  |
Note: This is the text of Eddy’s inaugural lecture at the Pontifical Academy of Sciences. You can follow the slides matching this presentation by opening the PowerPoint presentation immediately following this text.
EVO-DEVO: THE MERGING OF EVOLUTIONARY AND DEVELOPMENTAL BIOLOGY
EDWARD M. DE ROBERTIS
In the beginning of the 20th century, developmental biology was at the forefront of biology, but then declined and had a renaissance towards its end. The key to this revival were the techniques of molecular biology, which proved the great equalizer for all branches of biology. The fusion of molecular, developmental and evolutionary biology proved very fertile, and led to the birth of a new discipline, Evo-Devo. I would like to present a personal account on how this synthesis took place.
We will consider here three main points (slide 1).
1. How are the mechanisms of self-regulation of cell differentiation observed in animal development explained at the molecular level?
2. How were conserved ancestral gene networks common to all animals; which pattern the Antero-Posterior (A-P) and Dorso-Ventral (D-V) axes; used to generate the immense variety of animal forms?
3. How has the use of a common tool-kit of genes present in the ancestral animal genome channeled the outcomes of evolution through natural selection?
The main conclusion that emerges from these genomic, developmental and evolutionary studies is that all bilateral animals; which comprise 30 of the 34 extant phyla; arose through gene mutation, duplication or deletion of the genome of a complex common ancestor, the Urbilateria (Ur: primeval; Bilateria: animals having bilateral symmetry).
1. Self-regulation of differentiating cell fields
1.1 Embryology at the forefront of biology
When biologists realized that it was necessary to take an experimental – rather than descriptive – approach to understand the mechanisms of development, embryology rapidly became the leading edge of biological studies. Embryos offer an excellent material for experimental biology (slide 2).
After fertilization, an amphibian egg; a large cell 1.2 mm or more in diameter; divides synchronously into 2, 4, 8, 16, 32, 64 and so on cells. At these early stages, cells are dedicated to sensing their position within the embryo by signaling to each other without differentiating into particular tissues. At the 10,000 cell stage, cells on the dorsal side start to invaginate to the interior of what at this point constitutes a blastula or hollow ball. The cells that involute will form the endoderm and mesoderm of the body, while cells that remain in the outside give rise to ectoderm. By the end of this process – called gastrulation – a vertebrate embryo with defined A-P and D-V axes and differentiated tissue types is formed.
The beginning of experimental embryology can be traced back to 1891, when Hans Driesch separated the first two cells of a sea urchin embryo and obtained two complete larvae. At the turn of the century, in 1901, Hans Spemann obtained amphibian twins by gently constricting embryos with fine ligatures of hair from his newborn daughter (slide 3). Much later, I found that identical twins can also be generated by simply bisecting an early embryo of the frog Xenopus laevis with a scalpel blade before gastrulation starts.
This tendency of the embryo to regenerate towards the whole is called self-regulation. This is not a property restricted to the early embryo. Most organs in the body start their development as “morphogenetic fields” that are able to self-regulate their differentiation (slide 4). This was discovered by Ross G. Harrison, who showed in 1918 that a circular region of flank mesoderm could induce the development of forelimbs when transplanted into a host embryo. When he cut this region in half, each half induced a limb. Not a half-limb, but rather a complete limb. From these transplantation experiments we learned that cells within the organism do not lead solitary lives, but are instead subsumed in larger fields of hundreds or thousands of cells that communicate to each other when to proliferate, differentiate, or die. We are only now beginning to understand the molecular mechanisms by which these cellular conversations take place.
1.2 Hans Spemann and embryonic induction
The way forward in the analysis of self-regulation of pattern came from a transplantation experiment carried out by a graduate student at Freiburg University, Hilde Mangold (slide 5). Under the direction of Spemann, she transplanted the dorsal lip of the blastopore, the region in which the involution of mesoderm starts, and introduced it into the opposite (ventral) side of a host embryo. With a gentle push, the embryonic fragments heal together almost miraculously, and two days later perfect Siamese (conjoined) twins are formed. Spemann called this dorsal region of the embryo the “organizer”.
Remarkably, the transplanted organizer cells themselves gave rise to notochord, yet were able to induce their neighboring cells to change their differentiation into dorsal tissues such as central nervous system (CNS), somite (muscle), and kidney (slide 6). Therefore, within the embryo groups of cells (called organizing centers) are able to instruct their neighbors the type of cell differentiations they should adopt.
Spemann was awarded the 1935 Nobel Prize for Physiology or Medicine for the discovery of embryonic induction by organizer tissue, which marked the apogee of experimental embryology. However, the isolation of the chemical substances responsible for embryonic induction proved impossible given the methods available of the time. After that, the genetics of Thomas Hunt Morgan became the pre-eminent biological discipline for most of the 20th century.
2. The ancestral A-P and D-V gene networks
2.1 Thomas Morgan, Edward Lewis and homeotic mutations
Morgan started his career as an embryologist. For example, he demonstrated that a 2-cell frog egg could self-regulate to form a whole embryo after killing one cell, but only when the dead cell was removed. He realized, however, that mechanistic progress using this type of experimental approach would be very difficult, and decided to study mutations in the fruit fly Drosophila melanogaster instead (slide 7). Together with his graduate student Calvin Bridges, in 1923 Morgan isolated a mutant, bithorax, which gave rise to four-winged flies (flies normally have only two wings). This mutant was to provide the key that made possible the molecular analysis of development.
In 1946, a young student at Caltech, Edward B. Lewis, initiated studies on the genetics of the bithorax locus, which continued until his passing in 2004 (slide 8). He found that the bithorax region patterned the thorax and abdomen of the fruit fly and contained several genes. When mutated, these genes caused homeotic transformations, i.e., the transformation of one region of the body into the likeness of another region. For example, the third thoracic segment may become transformed into the second thoracic, thus generating the four-winged flies.
Remarkably, Lewis noted that the arrangement of homeotic genes in the DNA followed the same order in which they regulated the A-P identity of abdominal segments. He designated this surprising organization colinearity. Lewis proposed that homeotic genes had repressed thoracic identity in a centipede-like ancestor, and that recent duplications of these genes had further elaborated the identity of each abdominal segment.
When molecular biology became practical, the race to clone a homeotic gene began in several laboratories. It culminated with the isolation of Antennapedia, a homeotic gene that can transform antenna into leg, independently by Scott and Kaufman, and by Garber and Gehring in 1983. Searching for the hypothetical recently duplicated genes of Lewis, they discovered that many Drosophila homeotic genes crossreacted with a short region of DNA. This conserved segment of nucleic acid, called the homeobox, was found to encode a DNA-binding domain of 60 amino acids, designated the homeodomain (slide
2.2 Hox genes in vertebrates
At that time I was a professor in the same department as Walter Gehring at the Biozentrum of the University of Basel, Switzerland, and we shared group meetings. We decided to collaborate to test whether homeobox genes might be present in vertebrates. (The experiment was conceived for the wrong reasons: the first expression studies by Garber had shown Antennapedia expression in the CNS, and we suspected it might encode a peptide hormone, which were known at the time to have been conserved between Hydra and mammals). On the first try we cloned a gene, now called HoxC-6, from a Xenopus laevis genomic library which crossreacted with Antennapedia and ultrabithorax (Carrasco et al., 1984). The sequence of the homeodomain was very similar to that of Antennapedia. Later gene knockout studies by Mario Capecchi and others showed that this gene, like the other 39 Hox genes, caused A-P homeotic transformations when mutated in the mouse. This was a good thing, because in our paper in the last sentence of the introduction I had written: “If the frog gene cloned here eventually turns out to have functions similar to those of the fruit fly genes, it would represent the first development-controlling gene identified in vertebrates”. And it was so.
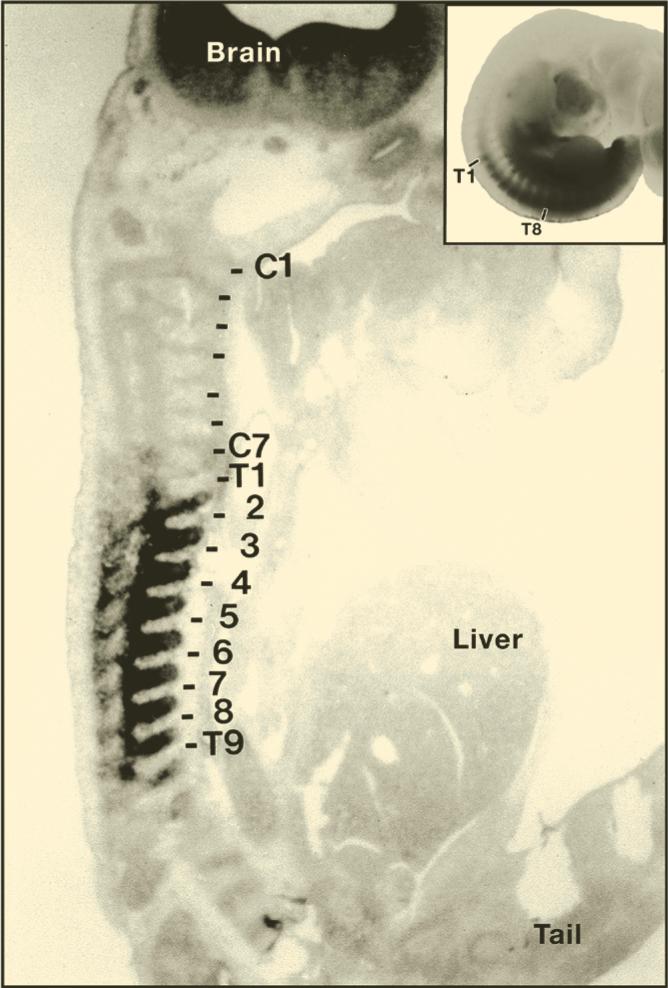
Vertebrate Hox genes are clustered in the genome. Work by other groups, mostly in mouse embryos, showed that vertebrate Hox gene expression in the body is colinear with their order in the DNA (slide 10). The homeobox sequences and overall organization of the vertebrate Hox gene complexes were conserved with those of Drosophila and other invertebrates. Therefore, Lewis’ hypothesis that homeotic genes were recently duplicated genes was not correct, yet provided the cornerstone for the new discipline of Evo-Devo. Edward Lewis received the Nobel Prize for Medicine or Physiology for his work on developmental genetics in 1995.
2.3. Whole-genome duplications in the vertebrate lineage
Many insects have eight or so Hox genes arranged in a single cluster. Amphioxus, a chordate closely related to the vertebrates, has a single cluster containing 14 Hox genes in a row. However, the situation is more complex in the vertebrates. This is because vertebrates underwent two rounds of whole-genome duplications at the beginning of their evolution. Thus, for each gene humans may have up to four copies. Many of our genes are now present as single copies, but this only indicates that the other three were lost. Gene loss is easily achieved over evolutionary time. Duplicated genes are retained when a duplicated copy acquires a specialized function that makes it beneficial for the survival of the species. These two genome-wide duplications were probably a crucial event in the remarkable evolutionary success of vertebrate animals.
Humans contain four Hox gene complexes, called HoxA through HoxD (slide 11). Each consists of about 100,000 base pairs of DNA and resulted from the duplication of an ancestral Hox complex containing 13 genes. However, instead of 13 x 4 = 52, humans retained a total of only 39 Hox genes. This is because some Hox genes were deleted. As will be discussed below, gene loss is an important force in shaping evolution.
The degree of conservation between these four mammalian Hox complexes and Drosophila is simply amazing. Not only homeobox sequences and colinearity of expression patterns were maintained, but even their regulation by an inhibitory microRNA (called infra-abdominal-4 in Drosophila and miR196 in humans) was conserved (slide 11).
This intricate genetic machinery that patterns the A-P axis could not have been assembled independently twice in Drosophila and vertebrates, let alone in all phyla. The only reasonable interpretation is that a Hox complex was already functional in Urbilateria and was inherited by its descendants (slide 12). The discovery of conserved Hox gene complexes led to the realization that the gene networks that control the A-P axis share deep historical homologies. Before the discovery of the homeobox we did not imagine that the mechanisms of development would be so similar between fruit flies and humans. It was a great surprise.
2.4. François Jacob’s symposium on Evolution and Development
In 1991, a landmark meeting was held in Crete. Organized, among others, by academicians Nicole Le Douarin and Fotis Kafatos, it was entitled “Evolution and Development”. Its topic had been specifically requested by François Jacob, who was retiring (slide 13). Jacob, a great geneticist, was very interested in evolution. In his excellent book, “The Possible and the Actual” (1982), Jacob explained why bringing these two separate fields together was important: “For it is during embryonic development that the instructions contained in the genetic program of an organism are expressed, that the genotype is converted into phenotype. It is mainly the requirements of embryonic development that, among all posible changes in genotype, screen the actual phenotypes.” The main argument of his book was that during evolution old components are retained and used again, comparing evolution to the work of a tinkerer or bricoleur. A tinkerer uses parts or materials that already exist to assemble objects having new purposes.
Jacob displayed great insight in bringing together developmental and evolutionary biologists as his swan’s song. The symposium took place at the perfect time, when the conservation of the Hox system was already understood in general outlines. The star of the meeting was paleontologist Stephen Jay Gould (slide 14). Wishing to learn more about evolution, I asked him to sit at my table during breakfast. Although he really wanted to read his newspaper in peace, I proved too eager and he reluctantly accepted. Gould recommended I should read two books. The first one was Gould’s own “Wonderful Life”, which told the story of the Cambrian explosion in the fossil record.
The Cambrian explosion refers to the remarkable finding that all the body plans (34 phyla) of animals that exist today appeared as fossils over a narrow period of time, between 535 to 525 million years ago. Before that time a long line of Precambrian ancestors must have existed, but they left very few or no adult bilaterian fossils (except for tracks and trails in the ocean floor dating to 630 million years ago). We do not know why the appearance of body plans occurred so suddenly, and many possibilities have been proposed (Valentine, 2004). For example, in the “snowball earth” scenario the diversification of body plans resulted from repeated bottlenecks of intense natural selection coinciding with several massive glaciations events that covered most of the earth between 750 and 550 million years ago. Even more mysterious than the sudden emergence of phyla, is the question of why no new animal body plans have evolved since then, for which we currently have no answers.
2.5. Geoffroy Saint-Hilaire and the unity of plan
The second book that Gould recommended was one by Toby Appel, on the historical debate that took place at the French Academy of Sciences between Georges Cuvier and Etienne Geoffroy Saint-Hilaire in 1830 (slide 14). Geoffroy held the view that a unity of plan existed among animals. In 1822, he dissected a lobster and placed it in an inverted position with respect to the ground. In this upside down orientation the lobster’s normally ventral nerve cord was located above the digestive tract, which in turn was placed above the heart. In his own words: “What was my surprise, and I add, my admiration, in perceiving an ordering that placed under my eyes all the organic systems of this lobster in the order in which they are arranged in mammals?”
Geoffroy went on to argue that there was a unity of plan, or design, among animals, so that the dorsal side of the vertebrates was homologous to the ventral side of the arthropods. For historians of science the Cuvier-Geoffroy debate was of great interest because it took place decades before Charles Darwin published his Origin of Species in 1859. For our own work, reading this book was crucial, because when a few years later we isolated Chordin, we were prepared. Chordin was a dorsal protein secreted by Spemann’s organizer that had a close homologue in the ventral side of the Drosophila early embryo.
At Jacob’s symposium I presented the first investigations from our laboratory on the chemical nature of embryonic induction by Spemann’s organizer. At that time, we had constructed libraries containing the genes expressed in dorsal lips manually dissected from the frog gastrula (slide 15). We had just isolated a gene expressed exclusively in organizer tissue called goosecoid. It encoded a DNA-binding protein, but we knew from Spemann’s work that embryonic induction required secreted factors able to change the differentiation of neighboring cells.
By continuing these explorations on the molecular nature of induction by organizer tissue, we isolated several secreted proteins such as Chordin, Frzb-1 and Cerberus, and other groups isolated Noggin, Follistatin and Dickkopf (De Robertis, 2006). Unexpectedly, all of these proteins turned out to function as antagonists of growth factors in the extracellular space. They prevent binding of growth factors to their receptors on the cell membrane, thus inhibiting signaling. Although we had hoped to isolate novel signaling growth factors from the organizer, what was discovered instead was that embryonic induction was mediated mainly through the secretion of a cocktail of inhibitory proteins.
2.6. Chordin, BMP and cell differentiation
Chordin proved to be the most informative of the organizer factors (slide 16). Transplanted organizers in which Chordin expression is inhibited lost all embryonic induction activity. Thus, Chordin is essential for organizer function. Chordin induces the differentiation of dorsal tissues (such as CNS or muscle) by binding to Bone Morphogenetic Proteins (BMPs), which normally cause the differentiation of ventral tissues (such as epidermis or blood). Two BMP genes are expressed in the ventral region of the embryo, and Chordin is secreted in prodigious amounts by dorsal cells. In principle, this would suffice to establish a gradient of BMP activity, yet by further investigating the system we discovered much more complexity.
Dorsal-ventral tissue differentiation results from a biochemical network of proteins secreted by the dorsal and ventral sides of the embryo (slide 17). For each action of the dorsal organizer there is a compensating reaction in the opposite side of the embryo. The expression of genes on the dorsal and the ventral sides are under opposite control, which explains in part the self-regulation phenomenon. The dorsal side also expresses BMPs, which when bound to Chordin are able to flow towards the ventral side. There, a protease called Tolloid specifically degrades Chordin, liberating BMPs for signaling through its cell surface receptors. The flow of Chordin and its cleavage by this protease are key steps in maintaining the self-regulating gradient of BMP activity. A number of additional secreted proteins (called Sizzled, Crossveinless-2, Twisted gastrulation and Crescent) function as feedback regulators, providing additional resilience to the D-V patterning system (De Robertis, 2009).
Remarkably, other investigators found that this basic biochemical network is also used to regulate cell differentiation along the D-V axis in the early embryos of many other organisms, such as Drosophila, beetles, spiders, hemichordates, amphioxus, zebrafish and chick (slide 18). This intricate molecular machinery is most unlikely to have evolved independently multiple times during evolution specifically to control D-V patterning. The reasonable conclusion is that the Chordin/BMP/Tolloid pathway patterned the dorsal-ventral axis of the last common bilaterian ancestor and was inherited by its descendants (slide 19).
The conservation of the Chordin/BMP/Tolloid system provided strong molecular support for the hypothesis of Geoffroy Saint-Hilaire that the mammalian and arthropod body plans are homologous. An inversion of the D-V axis occurred during evolution. The ventral side of the arthropods is equivalent to the dorsal side of the vertebrate, and the entire Chordin/BMP/Tolloid pathway was inverted. In both vertebrates and invertebrates, the CNS is formed where the gradient of BMP signaling is lowest. A unity of plan, both for the A-P and D-V axes, exists among animals.
3. A conserved gene tool-kit generates variety in evolution
3.1. Urbilateria had considerable regulatory complexity
These deep homologies in the way all embryos pattern their A-P and D-V axes are having a profound impact on current evolutionary thinking. One might argue that the power of natural selection of the fittest, working on chance mutations over immense periods of geological time, is per se sufficient to explain the variety of animal forms. In the absence of any constraints, competition in crowded ecosystems, particularly among closely related species, would lead to new and improved animal designs in the victorious species, through the creative force of natural selection. Ever more adapted generations would be formed because the invisible guiding hand of natural selection integrates useful mutational changes, forming ever fitter individuals and gradually generating new structures and species. On the other hand, what we are now learning is that a very important source of variation for specifying the arrangements of cells with respect to each other – which is what ultimately determines morphological change – resides in the ancestral developmental gene networks shared by all animals.
3.2 Eyes have a common origen
One might argue that while the Hox and Chordin/BMP gene networks are complex, they could have been used to pattern a very simple ancestral animal. However, there are reasons to think that Urbilateria was anatomically complex. One such reason is provided by the ancestral eye structures (slide 20).
An important problem in evolution is whether adaptations arise through homology or convergence. Homology means that two structures are derived from an ancestral one present in a common ancestor. An example of homology could be the hoof of a horse and the middle digit of the ancestors from which it evolved. Convergence occurs when similar solutions are reached to resolve common functional needs. An example could be the wings of various animal groups, which evolved at very different times but represent similar solutions to a functional requirement. Distinguishing between homology and convergence in evolution can be very difficult. Now molecular biology gives us a historical record of how evolution took place. In the case of animal eyes, conventional wisdom was that animal eyes had arisen independently 40 to 60 times through convergent evolution to fulfill the need for vision.
In 1994 Walter Gehring’s group isolated the eyeless gene from Drosophila and found it had homology to the mammalian Pax6 homeobox gene. In the mouse, mutations in Pax6 caused the small eye phenotype. In humans, the Aniridia gene corresponded to Pax6. When mouse Pax6 was artificially expressed in the antenna or leg precursors of Drosophila embryos (slide 21), it caused the formation of ectopic eyes (Gehring, 1998). Of course, these were Drosophila eyes, not mouse ones. In the reciprocal experiment, overexpression of Drosophila eyeless/Pax6 induced eyes in microinjected frog embryos. The eyes of the clam Pecten, and even those of jellyfish, also express Pax6. The reasonable conclusion is that all eyes are derived from an ancestral eye that expressed Pax6.
3.3 The urbilaterian CNS was anatomically elaborate
One might argue that the eye of Urbilateria could have been a very simple photoreceptor cell. However, this does not seem to be the case. We now have a very detailed understanding of the molecular switches (called transcription factors) that control the differentiation of the different neurons of the retina, which derives from the forebrain. The morphology of mammalian and Musca domestica eyes had been described in loving detail by Santiago Ramón y Cajal (slide 22). In 1915, he noted that by simply displacing the cell body (soma) of two neurons in Musca, leaving the cell projections and synaptic connections in place, the entire arrangements of intricate neural connections was maintained, with only small variations, between flies and humans. Recent studies have shown that the transcription factors expressed by various mammalian retinal neurons (photoreceptors, bipolar, and retinal ganglion cells) are replaced in the predicted corresponding fly neurons by their Drosophila homologues genes. This has provided molecular confirmation for Cajal’s homologies, which he had predicted from pure morphology (Sanes and Zipursky, 2010).
Extensive conservations in “molecular fingerprints” of particular combinations of transcription factors have also been found between vertebrate and Drosophila nerve cord neurons. In addition, brain hypothalamic neurosecretory cells express the same combinations of transcription factors as their corresponding Drosophila or annelid counterparts, which are located within the CNS region traversed by the mouth in protostomes. These neurosecretory peptides, important for sensing and signaling the availability of food, are expressed in the infundibulum of the mammalian brain, through which the gut probably traversed in our hypothetical ancestors (Tessmar-Raible et al., 2007). Thus, Urbilateria had a CNS, including eyes, that was sophisticated both from molecular and anatomical standpoints (slide 23). Before this stage was reached, a long line of Precambrian ancestors must have existed, in which their brains, neural circuits, and eyes were gradually perfected.
3.4 Animals share a conserved genomic tool-kit
Until recently the history of animal life on earth had to be deduced from the fossil record. Rapid advances in DNA sequencing have now made available entire sequenced genomes from multiple animal phyla. Because the genetic code arose only once, evolutionary studies are now less dependent on paleontology. We will be able to reconstruct the history of life on earth, registered in the language of DNA, with a degree of precision that seemed impossible only a decade ago (slide 24). For those interested on how animal evolution actually took place, comparative genomics offers the best of times.
The most important lesson we have learned so far from genome sequences is that animals from the most diverse phyla share a common ancestral tool-kit of genes (De Robertis, 2008). In particular, all the signaling pathways used by cells to communicate with each other – and therefore to regulate their anatomical position with respect with each other in the body – were already present in pre-bilaterian such as cnidarians (sea anemones, medusae and Hydra). Therefore, evolutionary changes resulted from the shuffling of a full ancestral set of genes, rather than from the introduction of new genetic mechanisms from scratch. There was remarkably little biochemical novelty during animal evolution.
3.5 Adaptive mutations
DNA sequencing has given us the opportunity of identifying the adaptive mutations that were actually selected during the evolution of animal populations in nature. The main types of variations on which natural selection acted to select the adaptive ones were: cis-regulatory mutations, structural gene mutations, gene duplications and gene deletions.
Cis-regulatory mutations are those found in the regulatory regions – called enhancers – located in cis (in the same DNA molecule) near genes. Enhancers regulate in which tissues genes are expressed. Enhancer DNA sequences provide binding sites for combinations of transcription factors that turn genes on and off. By changing the tissue or region in which a gene is expressed, morphological change can be generated. For example, crustaceans such as shrimp and lobsters evolved a considerable diversity of feeding appendages; it has been shown that these changes repeatedly correlated with independent shifts in the border of expression of Hox genes. New enhancers can be readily generated by bringing together combinations of DNA binding sites. They can also be easily lost without paying a large penalty, because the protein encoded by the gene remains and can still be expressed in other tissues under the control of the remaining enhancer elements. Mutations in tissue-specific enhancers are a major source of variations in evolution (Carroll, 2005). However, because enhancers are not highly conserved in sequence, their mutations are rarely detected by automatic sequence comparisons.
Structural mutations affect the sequence of the proteins encoded by genes. Interestingly, adaptive changes many times result from selection of mutations in the same gene. Melanism can be a useful adaptation. Melanic leopards, jaguars, mice, birds and lizards all arose from amino acid changes that increased the activity of the Melanocortin-1 receptor (Haekstra and Coyne, 2007). Conversely, decreased activity of this receptor is seen in yellow Labradors and human redheads. Thus, natural selection chooses the same solutions repeatedly.
Gene duplications are very powerful source of evolutionary variation because the duplicated gene can be used to fulfill new functions without loss of the original gene (Ohno, 1970). For the molecular biologist they offer the additional advantage that the duplication – or the deletion – of an entire gene is easily recognized when comparing genomic DNA sequences, thus facilitating the reconstruction of the history of animals.
Gene deletions are a very effective, although generally underappreciated, source of adaptation. Many cave animals – such as salamanders, shrimp and fish – adapt to their new troglodyte environment by losing their eyes and skin pigment. In the case of Mexican Tetra fish, their entrapment in subterraneous caves has led to deletion events in the ocular and cutaneous albinism gene-2 that occurred independently in different populations (Protas et al., 2006). Natural selection tends to choose mutations in the same genes over and over again. Although gene deletions are an effective way of rapidly adapting to changes in the environment, this is achieved at the expense of limiting future evolutionary flexibility.
3.6 Gene losses in the ancestral tool-kit
There are 30 bilateral animal phyla with distinct body plans, which can be classified in two branches. In the protostomes (mouth-first), the mouth is formed near the blastopore – these animals include most invertebrates. In the deuterostomes (mouth-second), the blastopore gives rise to the anus and the mouth is perforated secondarily – these animals include the phylum Chordata to which we belong. For example, if a gene is found both in fruit flies and in humans, it was also present in their last common ancestor, Urbilateria, as well. Similarly, if a gene is found both in pre-bilaterian animals such as sea anemones as well as in humans, it follows that this gene was also present in Urbilateria.
The role of gene loss in the evolution of Phyla has been highlighted by the sequencing of a sea anemone genome (slide 24). The bilaterian lineage separated from cnidarians, at least 650 million years ago, from a common animal ancestor designated Ureumetazoa. About 2.5% of sea anemone genes are not present in any higher animals but, interestingly, have homologues in fungi and plants. The human genome contains twenty-plus genes of the Wnt family of growth factors. These can be arranged into 13 subfamilies according to their sequence. The sea anemone has 12 Wnt genes, each corresponding to one of the human subfamilies. (Kusserow et al., 2005). Therefore, Urbilateria had genes corresponding to at least 12 Wnt subfamilies. Sequencing of the nematode C. elegans showed that it has a grand total of five Wnts; the Drosophila genome contains only seven. Thus, our human lineage retained most of the ancestral Wnt genes, while worms and fruit flies lost a great many. There are also examples in the opposite direction, in which humans have lost genes present in other vertebrates such as fish, frog or chick. Comparative genomics indicates that gene losses, as well as duplications, may have played an important role in the evolution of body plans.
3.7 Historical constraints in animal evolution
A key question in Evo-Devo is to what degree the deep homologies in embryonic patterning networks have channeled the outcomes of evolution (slide 25). Many body plans that could have been excellent functional solutions might not exist in nature because they cannot be constructed unless they are compatible with the developmental networks that control the blueprint of animal body form. The respective contributions of functional needs and structural constraints is of great interest in evolutionary biology (Gould, 2002). Paraphrasing François Jacob, not all that is possible finds its way into the actual animal world.
The deep homologies in the developmental tool-kit seem likely to have constrained animal evolution by natural selection. Constraints resulting from the obligatory use of these ancestral patterning networks should not be considered a negative influence. On the contrary, mutations in these gene networks may have been a positive influence that channeled effective adaptation responses to the strictures of natural selection. Adaptation tends to follow the channel of least resistance to ensure survival of the species and it seems likely that modifications in developmental networks have been used repeatedly to resolve related functional needs. Many anatomical structures now considered to result from convergent evolution may turn out to result from the deep homologies in the genetic structure of all animals. Evolution of animal forms involved tinkering with the conserved A-P, D-V, and other developmental gene networks.
3.8. Open questions in Evo-Devo.
Three directions will be particularly important for the young discipline of Evo-Devo (slide 26):
First, the reconstruction of the ancestral genetic tool-kit from which all animals were built should be a priority. This is at present a bioinformatic computing challenge. Many complete genome sequences are available already. Ideally the DNA of at least one species for each one of the 34 phyla should be completed. The ancestral tool-kit of yeasts has been determined and has proven interesting. Several groups are close to assembling an ancestral mammalian genome. Reconstructing the hypothetical genome of our urbilaterian ancestors will be very informative concerning the origin of body plans – particularly with respect to the role played by gene duplications and deletions during evolution.
Second, retracing the adaptive mutations that caused the actual anatomical changes selected by natural selection is another priority. Biology is a historical science, and it will be fascinating to unravel the successive molecular steps by which we evolved into our present human condition.
Third, determining how cells read their positional information in the embryo and adult tissues within self-regulating fields of cells will have both medical and evolutionary implications. In the organism, cells receive a multitude of signals that must be integrated and transformed into well-defined cell behaviors. These responses include cell division, differentiation and death, and are ultimately the determinants of morphological change.
CONCLUSION
The merging of Evolution and Development at the end of the 20th century has already provided important insights into how animals evolved an immense variety of body forms. The astonishing realization that has already emerged from Evo-Devo is that all animal life on earth evolved by differential use of the same ancestral tool-kit (slide 27). A crucial role was played by variations in ancestral developmental gene networks that are hard-wired within our DNA.
BIBLIOGRAPHY
Appel, T.A. (1987). The Cuvier-Geoffroy Debate. Oxford University Press, Oxford.
Cajal, S.R. and Sanchez, D. (1915). Contribución al conocimiento de los centros nerviosos de los insectos. Trab. Lab. Invest. Biol. 13, 1-167.
Carrasco, A.E., McGinnis, W., Gehring, W.J. and De Robertis, E.M. (1984). Cloning of a Xenopus laevis gene expressed during early embryogenesis that codes for a peptide region homologous to Drosophila homeotic genes: implications for vertebrate development. Cell 37, 409-414.
Carroll, S. (2005). Endless Forms Most Beautiful: The New Science of Evo-Devo. W.W. Norton & Co., Inc., New York.
Darwin, C. (1859). On the Origin of Species by Means of Natural Selection, or Preservation of Favored Races in the Struggle for Life. Murray, London.
De Robertis, E.M. (2006). Spemann’s organizer and self-regulation in amphibian embryos. Nat. Rev. Mol. Cell Biol. 7, 296-302.
De Robertis, E.M. (2008). Evo-Devo: Variations on Ancestral themes. Cell 132, 185-195.
De Robertis, E.M. (2009). Spemann’s organizer and the self-regulation of embryonic fields. Mech. Dev. 126, 925-941.
De Robertis, E.M. and Sasai, Y. (1996). A common plan for dorso-ventral patterning in Bilateria. Nature 380, 37-40.
Gehring, W.J. (1998). Master Control Genes in Development and Evolution: The Homeobox Story. Yale Univ. Press, New Haven.
Geoffroy Saint-Hilaire, E. (1822). Considérations générales sur la vertèbre. Mém. Mus. Hist. Nat. 9, 89-119.
Gould, S.J. (1989). Wonderful Life. W.W. Norton & Company, New York.
Gould, S.J. (2002). The Structure of Evolutionary Theory. Harvard University Press, Cambridge, Massachusetts. Chapter 10.
Hamburger, V. (1988). The Heritage of Experimental Embryology. Oxford University Press, New York.
Hoekstra, H.E. and Coyne, J.A. (2007). The locus of evolution: Evo Devo and the genetics of adaptation. Evolution Int. J. Org. Evolution 61, 995-1016.
Kusserow, A., Pang, K., Sturm, C., Hrouda, M., Lentfer, J., Schmidt, H.A., Technau, U., von Haeseler, A., Hobmayer, B., Martindale, M.Q. and Holstein, T.W. (2005). Unexpected complexity of the Wnt gene family in a sea anemone. Nature 433, 156-160.
Jacob, F. (1982). The Possible and the Actual. University of Washington Press, Seattle.
Monod, J. (1971). Chance & Necessity. Alfred A. Knopf, New York.
Ohno, S. (1970). Evolution by Gene Duplication. Springer-Verlag, Heidelberg.
Protas, M.E., Hersey, C., Kochanek, D., Zhous, Y., Wilkens, H., Jeffery, W.R., Zon, L.I., Borowsky, R. and Tabin, C.J. (2006). Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat. Gen. 38, 107-111.
Ratzinger, J. (1995). In the Beginning. B. Eerdmans Publishing Co., Michigan.
Sanes, J.R. and Zipursky, S.L. (2010). Design principles of insects and vertebrate visual systems. Neuron 66, 15-36.
Tessmar-Raible, K., Raible, F., Christodoulou, F., Guy, K., Rembold, M., Hausen, H. and Arendt, D. (2007). Conserved sensory-neurosecretory cell types in Annelid and fish forebrain: insights into Hypothalamus evolution. Cell 129, 1389-1400.
Valentine, J.W. (2004). On the origin of Phyla. The Univ. of Chicago Press, Chicago.
| The Powerpoint presentation for this talk can be accessed by opening the file beside this text. |  |
The ancestry of segmentation
E. M. De Robertis
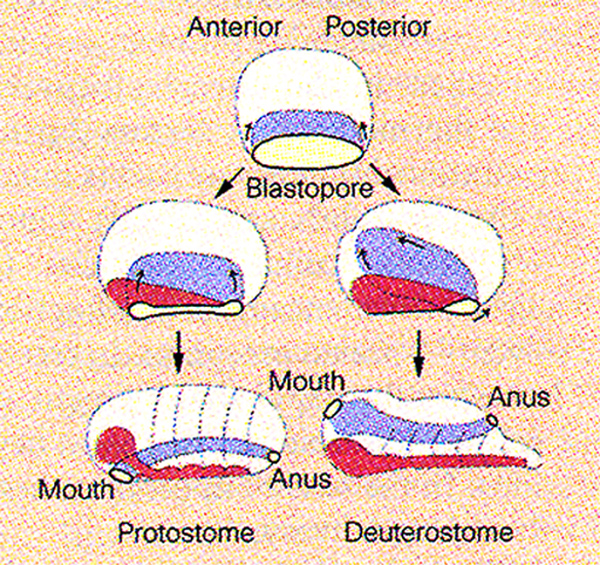
Evo-Devo: Variations on Ancestral Themes
E. M. De Robertis
Most animals evolved from a common ancestor, Urbilateria, which already had in place the developmental genetic networks for shaping body plans. Comparative genomics has revealed rather unexpectedly that many of the genes present in bilaterian animal ancestors were lost by individual phyla during evolution. Reconstruction of the archetypal developmental genomic tool-kit present in Urbilateria will help to elucidate the contribution of gene loss and developmental constraints to the evolution of animal body plans.
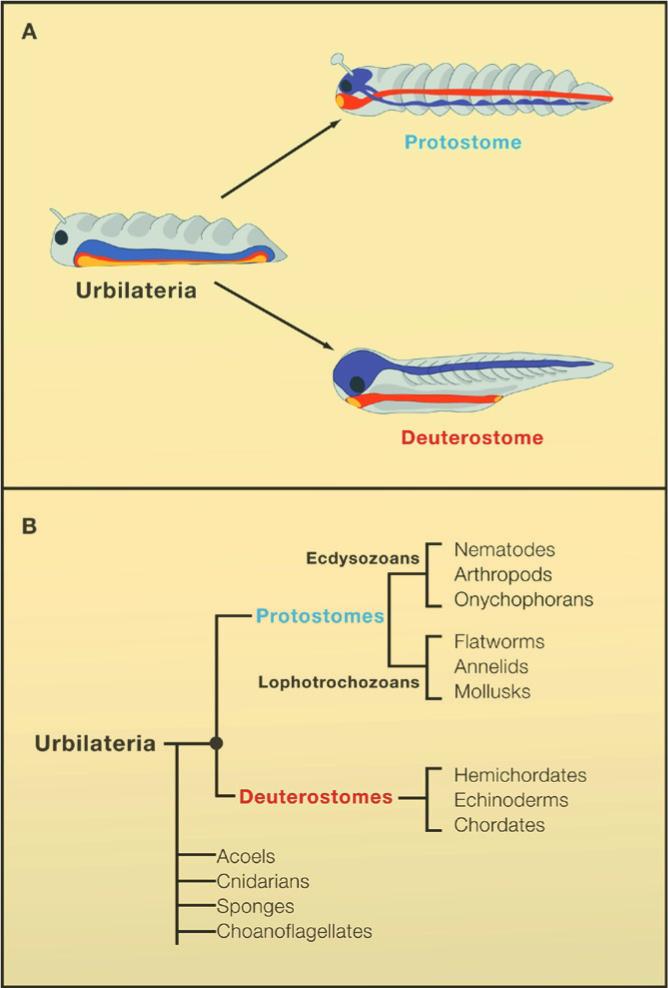 |
Figure 1. Evolutionary Relationships among Animals (A) Urbilateria is the archetypal animal that was the last common ancestor shared by protostomes and deuterostomes. The Urbilateria in this image is depicted as a segmented bottom-dwelling (benthic) animal with eyes, central nervous system, a small appendage, and an open slit-like blastopore. Endoderm is shown in red, central nervous system in dark blue, and surface ectoderm in light blue. (B) The new animal phylogeny, showing that cnidarians are basal to bilateria and that protostomes are divided into two branches, the molting Ecdysozoans and the nonmolting Lophotrochozoans. |
|
Figure 2. Hox Complexes of Drosophila and Mammals The Hox complex has been duplicated twice in mammalian genomes and comprises 39 genes. Note that microRNA genes, which inhibit translation of more anterior Hox mRNAs, have been conserved between Drosophila and humans. |
|
|
Figure 3. Posterior Repression of Hox-C6 mRN Translation in the Mouse Embryo Translation of Hox-C6 mRNA is seen in eight thoracic segments of the day 13 mouse embryo but blocked in the tail region, probably through the action of microRNAs. The inset shows that Hox-C6 mRNA is expressed all the way to the tip of the tail (using a Hox-C6-lacZ gene fusion). Note that the anterior border of expression of the Hox-C6 protein starts in the posterior half of the T1 segment, indicating that the sclerotome has already resegmented (G. Oliver and E.M.D.R., unpublished data; for methods see Oliver et al., 1988). |
|
|
Figure 4. The Conserved Chordin-BMP Signaling Network Although the Chordin-BMP signaling network is conserved, there has been a D-V axis inversion from Drosophila to Xenopus. (A) In Xenopus, Chordin is expressed on the dorsal side and BMP4 at the opposite ventral pole (image courtesy of Hojoon X. Lee). (B) In Drosophila, Dpp is dorsal (blue) and Sog is ventral (in brown) in the ectoderm (Image courtesy of Ethan Bier and reproduced from François et al., 1994, Genes Dev. 8, 2602−2616, with permission from Cold Spring Harbor Laboratory Press, copyright 1994). (C) A network of conserved secreted proteins mediates D-V body patterning in Xenopus and Drosophila. |
|
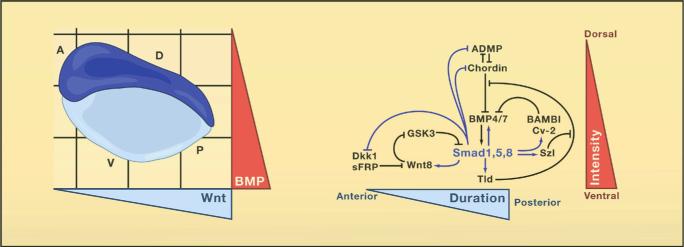 |
Figure 5. A-P and D-V Integration in the Embryonic Morphogenetic Field Shown is a model of the embryonic morphogenetic field in which a Cartesian System of A-P (Wnt) and D-V (BMP) gradients are integrated at the level of phosphorylation of the transcription factors Smads 1, 5, and 8. Note that in this self-regulating model the BMP gradient provides the intensity, but the Wnt gradient controls the duration of the Smad 1, 5, 8, signal. Direct protein-protein interactions are shown in black, and transcriptional activity of Smads 1, 5, and 8 are in blue. |
The molecular ancestry of segmentation mechanisms
E. M. De Robertis
Proc. Natl. Acad. Sci USA 105, 16411-16412 (2008)

| A common plan for dorsoventral patterning
E. M. De Robertis and Yoshiki Sasai |
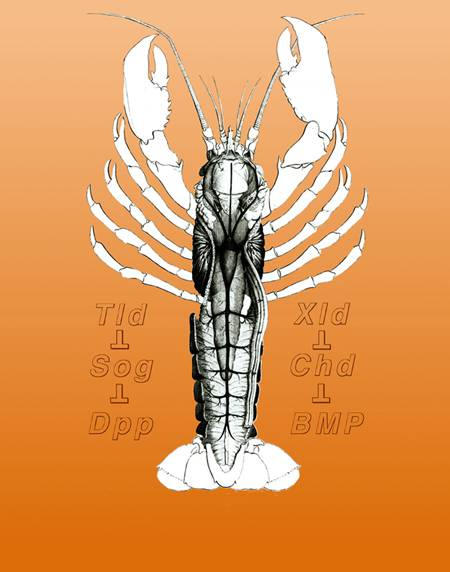 |
 |
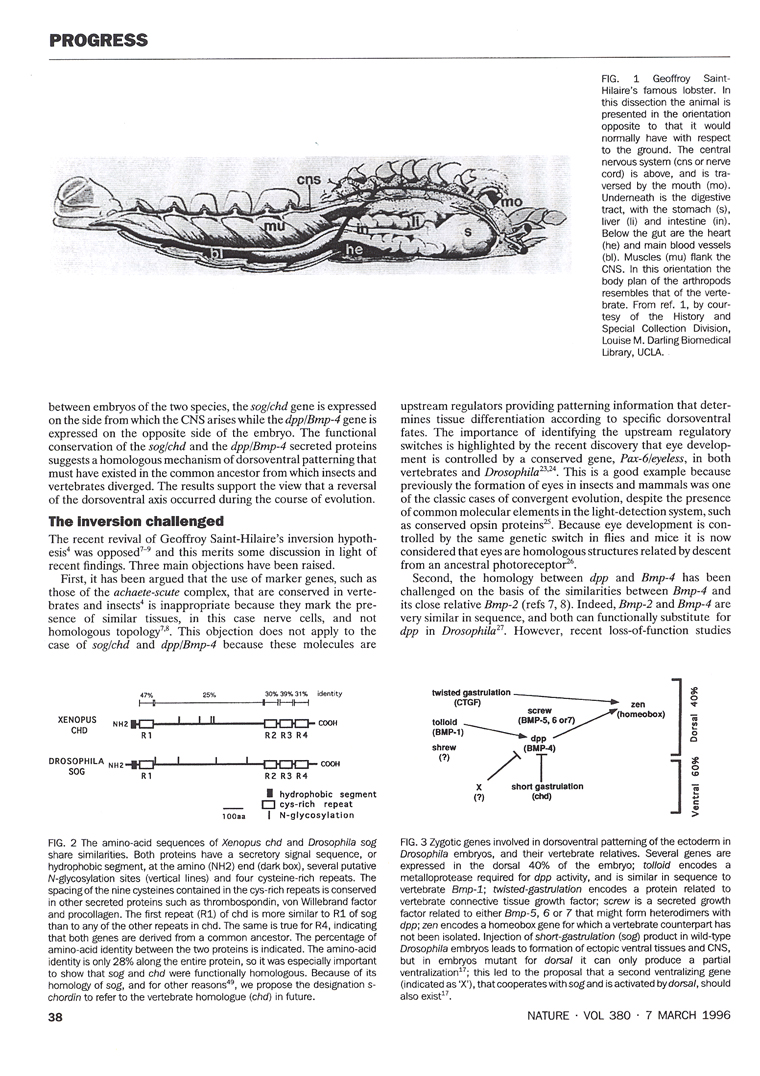 |
 |
 |
Homeobox genes and the vertebrate body plan
E. M. De Robertis, G. Oliver and C.V.E. Wright
Scientific American 263, 46-52 (1990)
|
|
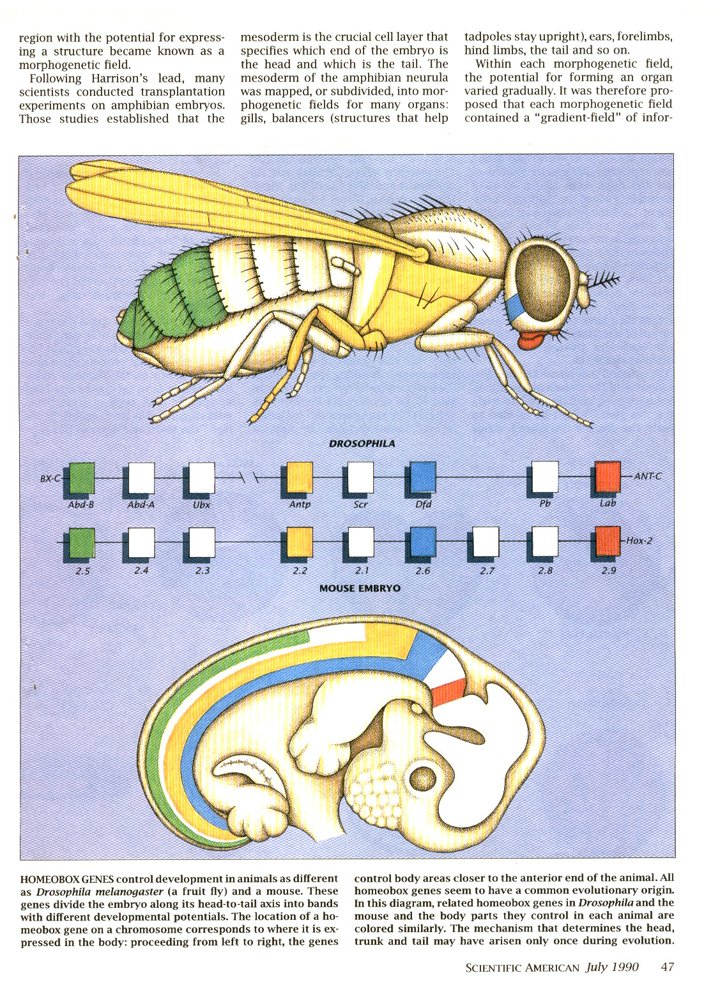 |
 |
 |
 |
 |
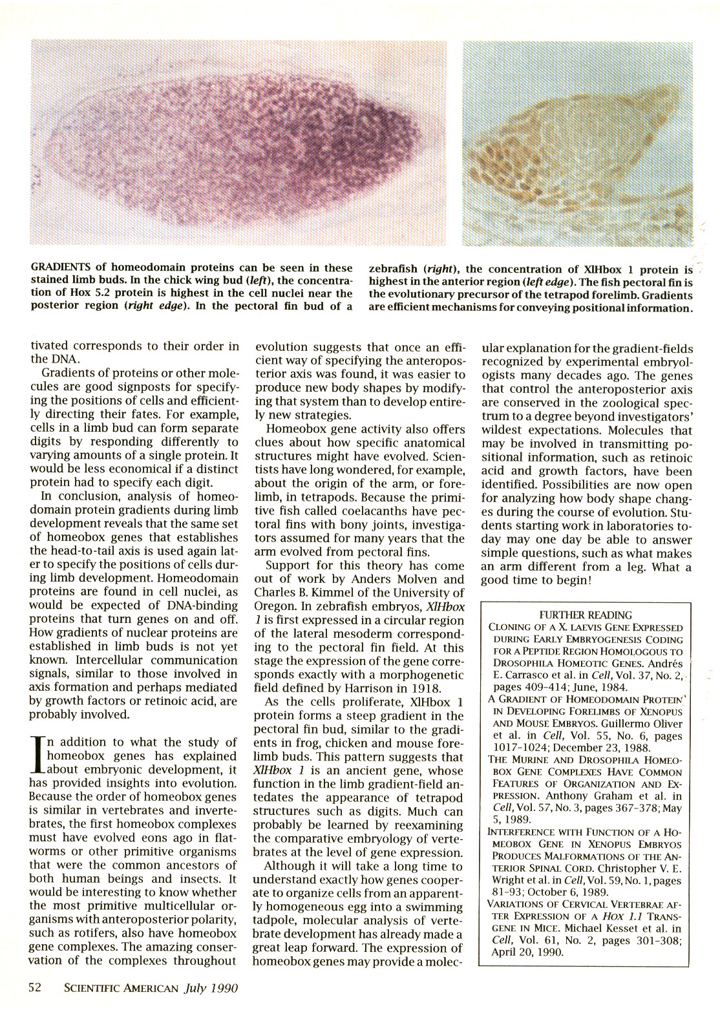 |