 |
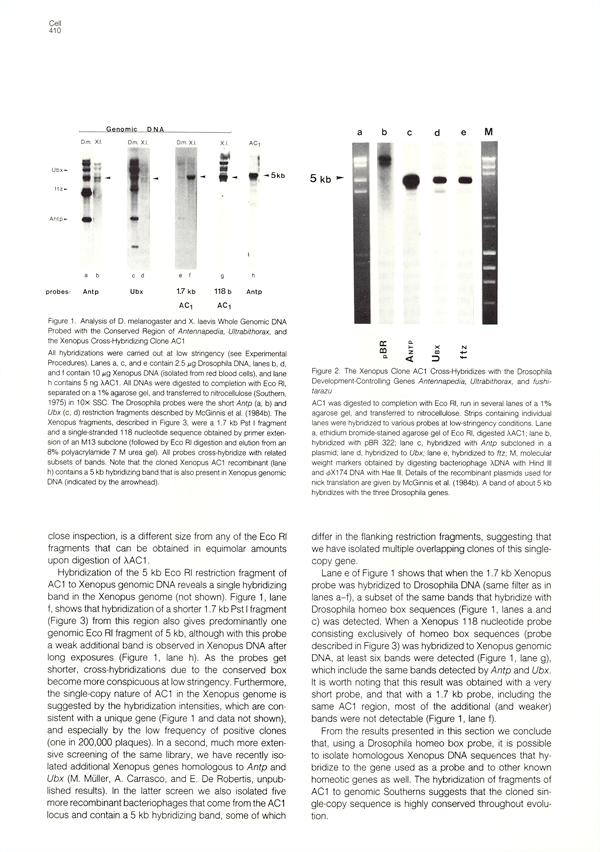 |
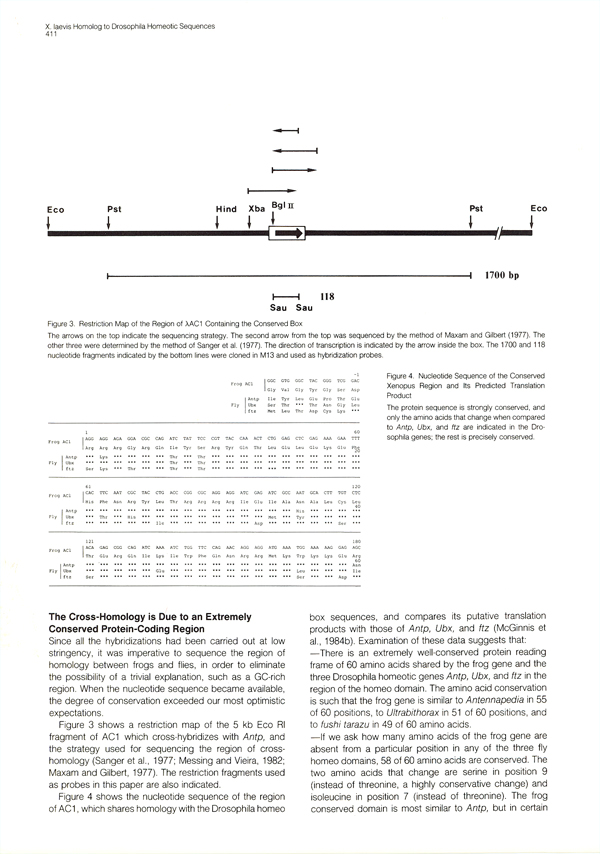 |
 |
 |
 |
Complementary homeoprotein gradients in developing limb buds
Guillermo Oliver, N. Sidell, W. Fiske, C. Haenzmann, T. Mohandas, R.S. Sparkes and E.M. De Robertis
A new human homeo box-containing gene designated -5.2 was cloned and mapped to human chromosome 2. This homeo box is related in sequence to Abdominal-B, a Drosophila homeotic gene that specifies identity of posterior segments. An antibody probe was made using a human -5.2 fusion protein and was found to stain posterior regions of mouse, chicken, and Xenopus embryos. Unexpectedly, when the distribution of -5.2 antigen was compared with that of X1Hbox 1 antigen, a non-overlapping and mutually exclusive pattern was detected (e.g., in developing limb buds, intestine, and somites). Regions expressing -5.2 do not express X1Hbox 1 protein, and vice versa. -5.2 antigen is detected strongly in developing fore- and hindlimb buds, where it forms a gradient of nuclear protein throughout most of the mesenchyme. This gradient is maximal in distal and posterior regions. -5.2 expression is activated in Xenopus limb regeneration blastemas, as expected for any gene involved in pattern formation. As described previously, a gradient of X1Hbox 1 protein can be detected in the forelimb. The latter gradient has the opposite polarity to that of -5.2. i.e., maximal in anterior and proximal mesoderm. These two opposing gradients (and possibly others) could be involved in determining positional values in developing limb buds.
Differential utilization of the same reading frame in a Xenopus homeobox gene encodes two related proteins sharing the same DNA-binding specificity
Ken W.Y. Cho, J. Getz, Chris V.E. Wright, Andreas Fritz, Jane Hardwicke and E. M. De Robertis
Xenopus XlHbox 1 produces two transcripts during early development. One encodes a long open reading frame (ORF) and the other a short ORF sharing the same homeodomain, but differing by an 82 amino acid domain at the amino terminus. The long protein amino terminus is conserved with many other homeodomain proteins, and its absence from the short protein could have functional consequences. Some viral genes also utilize a single ORF to encode transcription factors of antagonistic functions. The overall organization of the homologous genes in frog and man is similar, supporting the notion that both transcripts are of functional significance. Studies on XlHbox 1 function show that the region common to the long and short proteins has a sequence-specific DNA-binding activity, and that microinjection of specific antibodies into embryos results in the loss of structures derived from cells normally expressing XlHbox 1.
Differential antero-posterior expression of two proteins encoded by a homeobox gene in Xenopus and mouse embryos
Guillermo Oliver, Chris V.E. Wright, Jane Hardwicke and E. M. De Robertis
The X.laevis XlHbox 1 gene uses two functional promoters to produce a short and a long protein, both containing the same homeodomain. In this report we use specific antibodies to localize both proteins in frog embryos. The antibodies also recognize the homologous proteins in mouse embryos. In both mammalian and amphibian embryos, expression of the long protein starts more posteriorly than that of the short protein. This difference in spatial expression applies to the nervous system, the segmented mesoderm and the internal organs. This suggests that each promoter from this gene has precisely restricted regions of expression along the anterior-posterior axis of the embryo. Because the long and short proteins share a common DNA-binding specificity but differ by an 82 amino acid domain, their differential distribution may have distinct developmental consequences.
A gradient of homeodomain protein in developing forelimbs of Xenopus and mouse embryos
Guillermo Oliver, Chris V.E. Wright, Jane Hardwicke and E. M. De Robertis
The expression of the homeodomain protein XIHbox 1 in developing Xenopus limbs was analyzed using specific antibodies. In the forelimb bud mesoderm XIHbox 1 shows a clear antero-posterior gradient that is strongest in the anterior and proximal region of the forelimb. Hindlimb bud mesoderm is devoid of XIHbox 1, indicating an early molecular difference between arm and leg. The innermost ectodermal cell layer is positive throughout the forelimb and hindlimb bud ectoderm, but no other areas of the skin. Similar results are obtained in developing mouse limbs, suggesting that XIHbox 1 participates in forelimb development in a variety of tetrapods. In early tadpoles analyzed at stages preceding limb bud formation, the lateral plate mesoderm is positive in the region corresponding to the earliest “field” of forelimb information, but not in the hindlimb field. These results suggest a molecular link between morphogenetic fields, gradients, and homeobox genes in vertebrate development.
Homeobox genes and the vertebrate body plan
E. M. De Robertis, Guillermo Oliver and Chris V.E. Wright
Scientific American 263, 46-52 (1990)
This family of related genes determines the shape of the body. It subdivides the embryo along the head-to-tail axis into fields of cells that eventually become limbs and other structures.
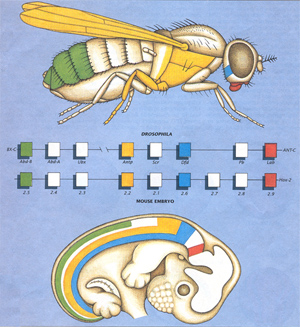
This figure, drawn by Guillermo Oliver, is of historical interest because
it was the first diagram of this kind. It has been modified many, many times.
Differential activation of Xenopus homeobox genes by mesoderm-inducing growth factors and retinoic acid
Ken W.Y. Cho and E.M. De Robertis
Genes Dev. 4, 1910-1916 (1990)
What is the nature of positional information during embryogenesis? By using Xenopus homeo box genes as anteroposterior (A-P) markers, we confirm the findings of others that mesoderm-inducing growth factors and retinoic acid (RA) can provide positional information along the axis of the body. Xenopus tissue culture-mesoderm-inducing factor (XTC-MIF) selectively activates an anteriorly expressed homeo box gene (XlHbox 1), while basic fibroblast growth factor (bFGF) activates selectively a posteriorly expressed homeo box gene (XlHbox 6). RA activates expression of the posterior gene XlHbox 6, but not of XlHbox 1. This activation, however, requires exposure to growth factors. The data suggest that growth factors and RA may cooperate with each other to provide positional information in vertebrates.
Homeotic transformations in the mouse induced by overexpression of a human 3.3
Beatriz C. Jegalian and E.M. De Robertis
A permanent transgenic mouse line was generated carrying 40 copies of the human 3.3 gene. The resulting mice express large amounts of 3.3 protein in posterior regions of the embryo where this homeodomain protein is normally not expressed. The transgene causes homeotic transformations of the skeleton, in particular the appearance of an extra pair of ribs in the lumbar region, transformation of the shape of posterior ribs into that of more anterior ones, and the joining of an additional pair of ribs to the sternum. The phenotype of this line resembles that obtained by the targeted loss-of-function mutation of 3.1 (Le Mouellic et al., 1992). In transient assays, the human 3.3 transgene leads to the formation of additional ribs in more posterior vertebrae as well.
Evo-Devo: variations on ancestral themes
E. M. De Robertis
Most animals evolved from a common ancestor, Urbilateria, which already had in place the developmental genetic networks for shaping body plans. Comparative genomics has revealed rather unexpectedly that many of the genes present in bilaterian animal ancestors were lost by individual phyla during evolution. Reconstruction of the archetypal developmental genomic tool-kit present in Urbilateria will help to elucidate the contribution of gene loss and developmental constraints to the evolution of animal body plans.